I want to create and fill a Silver layer based on parquet files in my bronze layer. Is there a simple way to create and populate the tables automatically.
 |
| Adding files to your Silver layer |
Solution
You can create a notebook for this and then call that notebook from your Synapse pipeline with some parameters (location, table name and keys). This allows you to for example loop through all your ingested source files from the bronze (raw/ingest) layer and then call this notebook for each file to add them to the Silver layer. We can also add the silver tables directly to the Lake database for easy querying later on.
Note: that this example is a technical source based Silver layer. So not realy cleansed, curated or conformed.
1) Create notebook
Go to the developer tab in Synapse and create a new Notebook. Give it a suitable name and make sure the language is PySpark. Sooner or later you want to test this Notebook, so attach it to a SparkPool. Optionally you can add a Markdown cell to explain this notebook.
2) Code cell 1: parameters
The first code cell is for the parameters that can be overridden by parameters from the Notebook activity in the pipeline. Toogle the parameters option to make is a parameter cell. For more details see our post about notebook parameters. For debugging within the notebook we used real values.
For this example everything (bronze and silver) is in the same container. So you might want to add more parameters to split those up. This example uses parquet files as a source. If you want for example CSV then you need to change the format in the mail code to fill the Spark Data Frame with data.
# path of the data lake container (bronze and silver for this example) data_lake_container = 'abfss://mysource@datalakesvb.dfs.core.windows.net' # The ingestion folder where your parquet file are located bronze_folder = 'Bronze' # The silver folder where your Delta Tables will be stored silver_folder = 'Silver' # The name of the table table_name = 'SalesOrderHeader' # The wildcard filter used within the bronze folder to find files source_wildcard = 'SalesOrderHeader*.parquet' # A comma separated string of one or more key columns (for the merge) key_columns_str = 'SalesOrderID'
 |
| Parameters |
3) Code cell 2: import modules and functions
The second code cell is for importing all required/useful modules. For this basic example we have only one import:
- DeltaTable.delta.tables
# Import modules from delta.tables import DeltaTable
 |
| Import Delta Table module |
3) Code cell 3: filling delta lake
Now the actual code for filling the delta lake tables with parquet files from the data lake. Note: code is very basic. It checks whether the Delta Lake table already exists. If not it creates the Delta Lake table and if it already exists it merges the new data into the existing table. If you have transactional data then you could also do an append instead of a merge.# Convert comma separated string with keys to array
key_columns = key_columns_str.split(',')
# Convert array with keys to where-clause for merge statement
conditions_list = [f"existing.{key}=updates.{key}" for key in key_columns]
# Determine path of source files from ingest layer
source_path = data_lake_container + '/' + bronze_folder + '/' + source_wildcard
# Determine path of Delta Lake Table
delta_table_path = data_lake_container + '/' + silver_folder + '/' + table_name
# Read file(s) in spark data frame
sdf = spark.read.format('parquet').option("recursiveFileLookup", "true").load(source_path)
# Check if the Delta Table exists
if (DeltaTable.isDeltaTable(spark, delta_table_path)):
print('Existing delta table')
# Read the existing Delta Table
delta_table = DeltaTable.forPath(spark, delta_table_path)
# Merge new data into existing table
delta_table.alias("existing").merge(
source = sdf.alias("updates"),
condition = " AND ".join(conditions_list)
).whenMatchedUpdateAll(
).whenNotMatchedInsertAll(
).execute()
# For transactions you could do an append instead of a merge
# sdf.write.format('delta').mode('append').save(delta_table_path)
else:
print('New delta table')
# Create new delta table with new data
sdf.write.format('delta').save(delta_table_path)
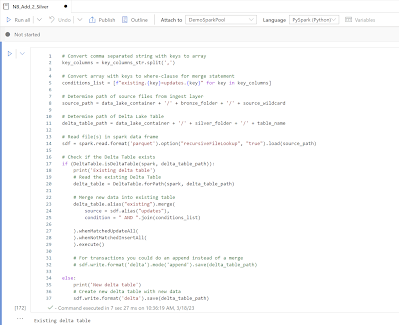 |
| Adding data to new or existing Delta Table |
4) Code cell 4: Adding Delta Table to Lake Database
The last step is optional, but very useful: adding the Delta Table to the Lake Database. This allows you to query the Delta Table by its name instead of its path in the Data Lake. Make sure you first add a Silver layer to that Lake database. See this post for more details (step 1).
# Adding the Delta Table to the Delta Database for easy querying in other notebooks or scripts within Synapse.
spark.sql(f'CREATE TABLE IF NOT EXISTS Silver.{table_name} USING DELTA LOCATION \'{delta_table_path}\'')
# Spark SQL version
# CREATE TABLE Silver.MyTable
# USING DELTA
# LOCATION 'abfss://yourcontainer@yourdatalake.dfs.core.windows.net/Silver/MyTable'
 |
| Adding Delta Table to Lake Database |
5) Creating Pipeline
Now it is time to loop through your ingested files and call this new Notebook for each file to create the Silver Layer Delta Tables. You have to provide values for all parameters in the notebook. Since you need the key column(s) of each table to do the merge you probably need to store these somewhere.
For the ingestion we often store the table/file names from each source that we want to download to the data lake in a meta data table. In this table we also store the key column(s) from each table.
Synapse doesn't retrieve the parameters from the Notebook. You have to add them manually as Base parameters in the Settings tab.
If you enter a column or set of columns for the key that are not unique you will get an error the second time you run (first time the merge is not used).
Cannot perform Merge as multiple source rows matched and attempted to modify the same target row in the Delta table in possibly conflicting ways. By SQL semantics of Merge, when multiple source rows match on the same target row, the result may be ambiguous as it is unclear which source row should be used to update or delete the matching target row. You can preprocess the source table to eliminate the possibility of multiple matches.
6) Result
Now you can run your pipeline and check whether the silver layer of you Lake database is populated with new tables. And you can create a new notebook with Spark SQL or PySpark to check the contents of the tables to see wether the Time Travel works.
Conclusions
In this post you learned how to create and populate a (source based) silver layer of your Lake House Delta Tables. An easy quick start for your lake house. If you have multiple sources with similar data then you should also consider creating a real cleansed, curated and conformed silver layer manually. In a later post we will show you some of those manual steps in Spark SQL or PySpark.
Special thanks to colleague Heleen Eisen for helping out with the PySpark.




